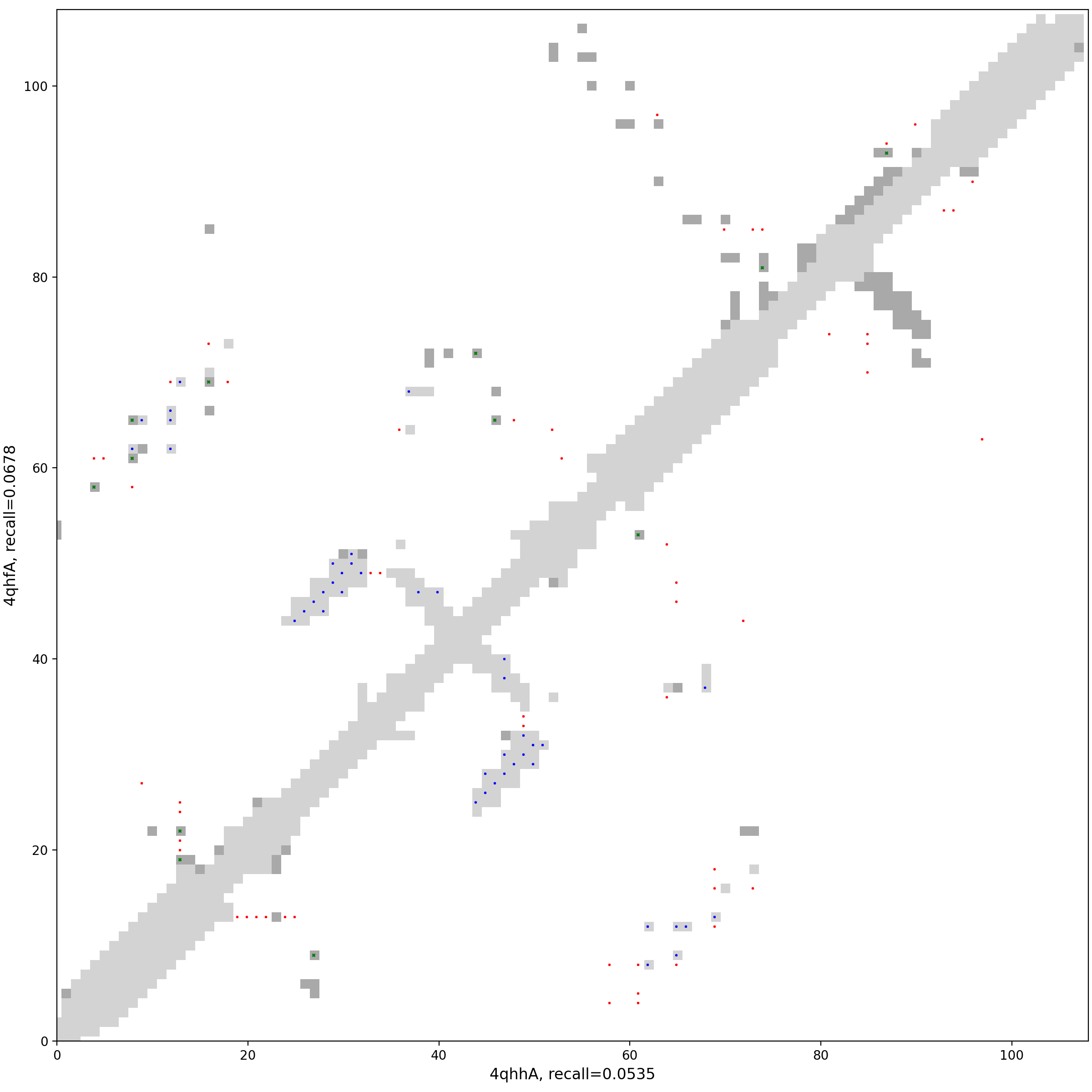
Figure 1: Two structures (Fold1 & Fold2)

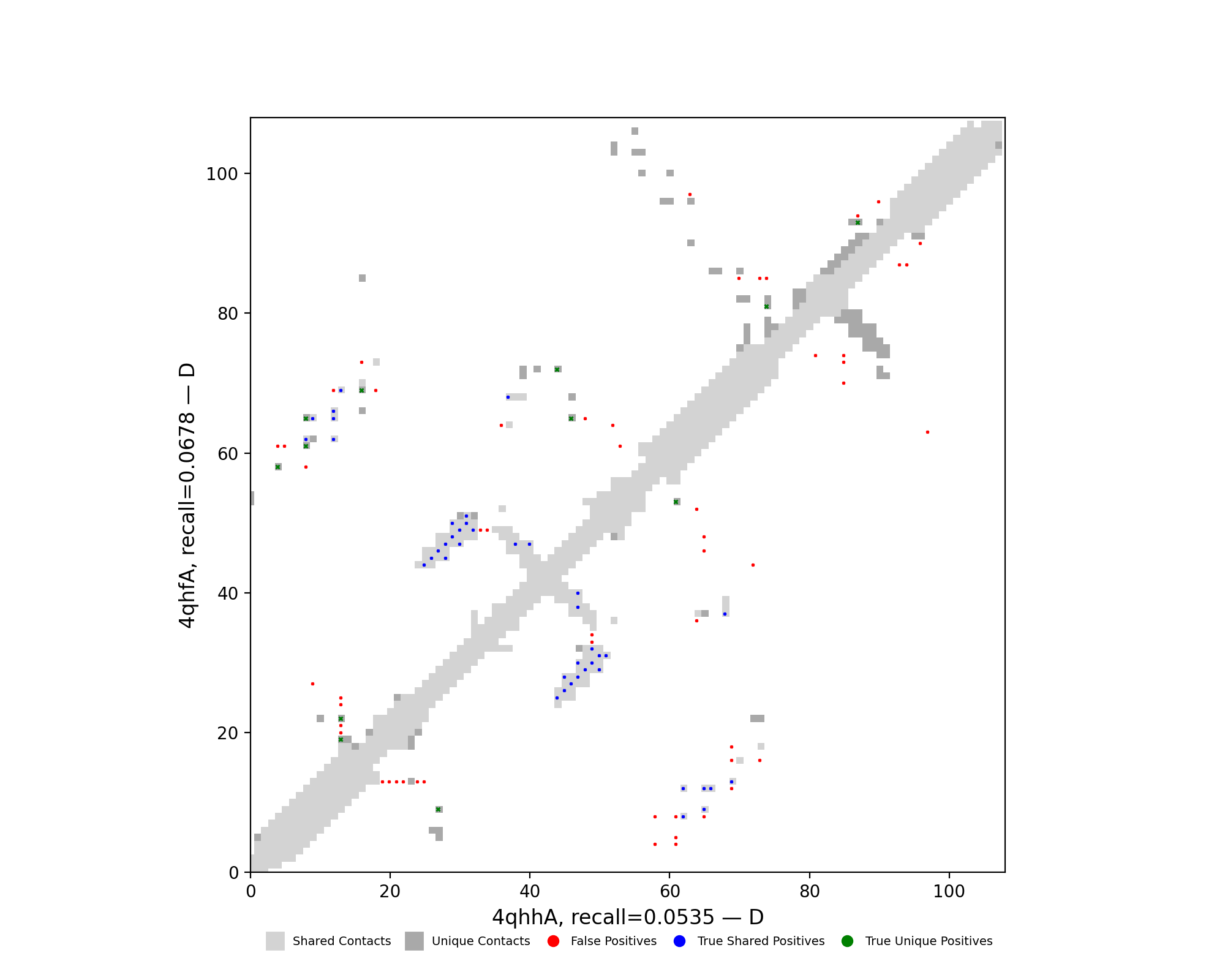
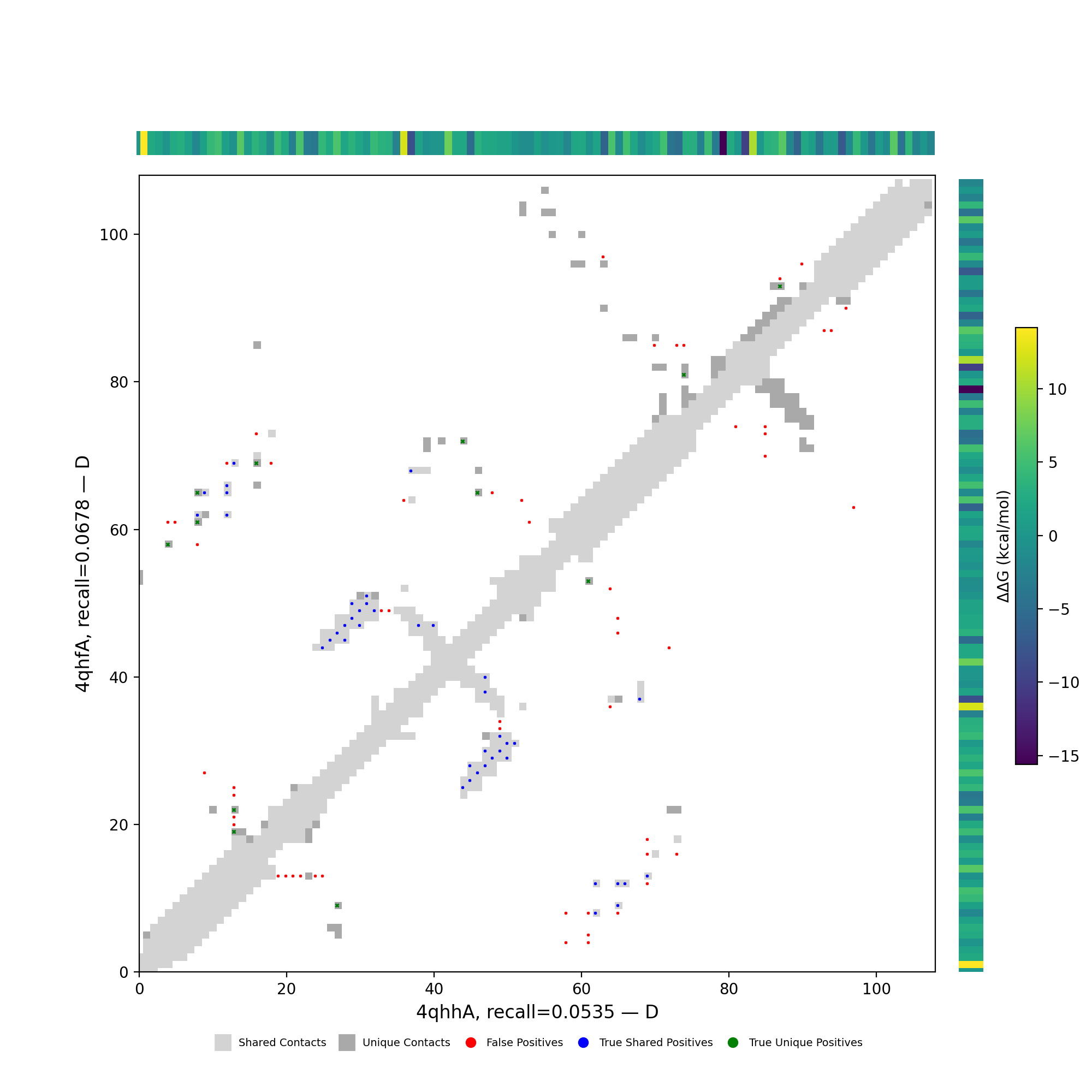
| cluster | n | neff | AF2_TM1 | AF2_TM2 | AF3_TM1 | AF3_TM2 | RE-MSAT-COM | RE-MSAT1 | RE-MSAT2 | ESM_SEQIDS | ESM_TM1_LIST | ESM_TM2_LIST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deep | - | - | 0.64 | 0.95 | 0.64 | 0.95 | 0.37 | 0.34 | 0.43 | - | - | - |
| 000 | - | - | 0.65 | 0.95 | 0.64 | 0.95 | 0.39 | 0.34 | 0.52 | 001; 001; 003; 003; 004; 004; 002; 006; 002; 006 | 0.65; 0.65; 0.63; 0.63; 0.62; 0.61; 0.55; 0.54; 0.54; 0.54 | 0.94; 0.94; 0.85; 0.85; 0.62; 0.69; 0.69; 0.71; 0.70; 0.71 |
| 001 | - | - | 0.34 | 0.34 | 0.26 | 0.25 | 0.0 | 0.0 | 0.0 | 002; 007; 002; 007; 010; 010; 001; 001; 004; 004 | 0.36; 0.36; 0.36; 0.36; 0.35; 0.35; 0.33; 0.33; 0.32; 0.32 | 0.36; 0.36; 0.36; 0.36; 0.36; 0.36; 0.33; 0.33; 0.31; 0.31 |
| 002 | - | - | 0.26 | 0.49 | 0.27 | 0.49 | 0.0 | 0.0 | 0.0 | 002; 002; 005; 005; 009; 009; 007; 007; 006; 006 | 0.48; 0.48; 0.46; 0.46; 0.41; 0.41; 0.38; 0.38; 0.36; 0.36 | 0.48; 0.48; 0.47; 0.47; 0.40; 0.40; 0.39; 0.39; 0.37; 0.37 |
| 003 | - | - | 0.46 | 0.53 | 0.45 | 0.49 | 0.04 | 0.04 | 0.03 | 006; 005; 006; 005; 001; 001; 008; 008; 002; 002 | 0.57; 0.57; 0.57; 0.57; 0.54; 0.54; 0.50; 0.50; 0.46; 0.46 | 0.69; 0.70; 0.69; 0.70; 0.66; 0.66; 0.69; 0.69; 0.59; 0.59 |
| 004 | - | - | 0.42 | 0.47 | 0.38 | 0.42 | 0.0 | 0.0 | 0.0 | 009; 009; 004; 003; 003; 004; 008; 008; 006; 006 | 0.55; 0.55; 0.54; 0.54; 0.54; 0.54; 0.48; 0.48; 0.45; 0.45 | 0.64; 0.64; 0.66; 0.66; 0.66; 0.66; 0.62; 0.62; 0.55; 0.55 |
| 005 | - | - | 0.39 | 0.38 | 0.42 | 0.4 | 0.0 | 0.0 | 0.0 | 001; 009; 009; 001; 006; 006; 003; 007; 003; 007 | 0.42; 0.42; 0.42; 0.42; 0.38; 0.38; 0.35; 0.35; 0.35; 0.35 | 0.42; 0.41; 0.41; 0.42; 0.38; 0.38; 0.32; 0.36; 0.32; 0.36 |
| 006 | - | - | 0.26 | 0.24 | 0.2 | 0.19 | 0.0 | 0.0 | 0.0 | 003; 006; 003; 006; 004; 004; 001; 002; 001; 002 | 0.30; 0.30; 0.30; 0.30; 0.29; 0.29; 0.28; 0.28; 0.28; 0.28 | 0.28; 0.28; 0.28; 0.28; 0.28; 0.28; 0.28; 0.26; 0.28; 0.26 |
| 007 | - | - | 0.57 | 0.66 | 0.45 | 0.49 | 0.07 | 0.05 | 0.08 | 008; 008; 001; 001; 006; 006; 002; 002; 004; 004 | 0.64; 0.64; 0.63; 0.63; 0.59; 0.59; 0.59; 0.59; 0.58; 0.58 | 0.82; 0.82; 0.80; 0.80; 0.57; 0.56; 0.58; 0.58; 0.55; 0.55 |
| 008 | - | - | 0.26 | 0.26 | 0.24 | 0.26 | 0.0 | 0.0 | 0.0 | 002; 003; 003; 002; 001; 001; 006; 006; 007; 008 | 0.30; 0.30; 0.30; 0.30; 0.26; 0.26; 0.24; 0.24; 0.24; 0.23 | 0.29; 0.29; 0.29; 0.29; 0.25; 0.25; 0.22; 0.22; 0.20; 0.19 |
| 009 | - | - | 0.3 | 0.3 | 0.19 | 0.17 | 0.0 | 0.0 | 0.0 | 008; 008; 002; 002; 007; 007; 001; 001; 009; 009 | 0.35; 0.35; 0.32; 0.32; 0.32; 0.32; 0.31; 0.31; 0.30; 0.30 | 0.34; 0.34; 0.32; 0.32; 0.31; 0.31; 0.31; 0.31; 0.30; 0.30 |
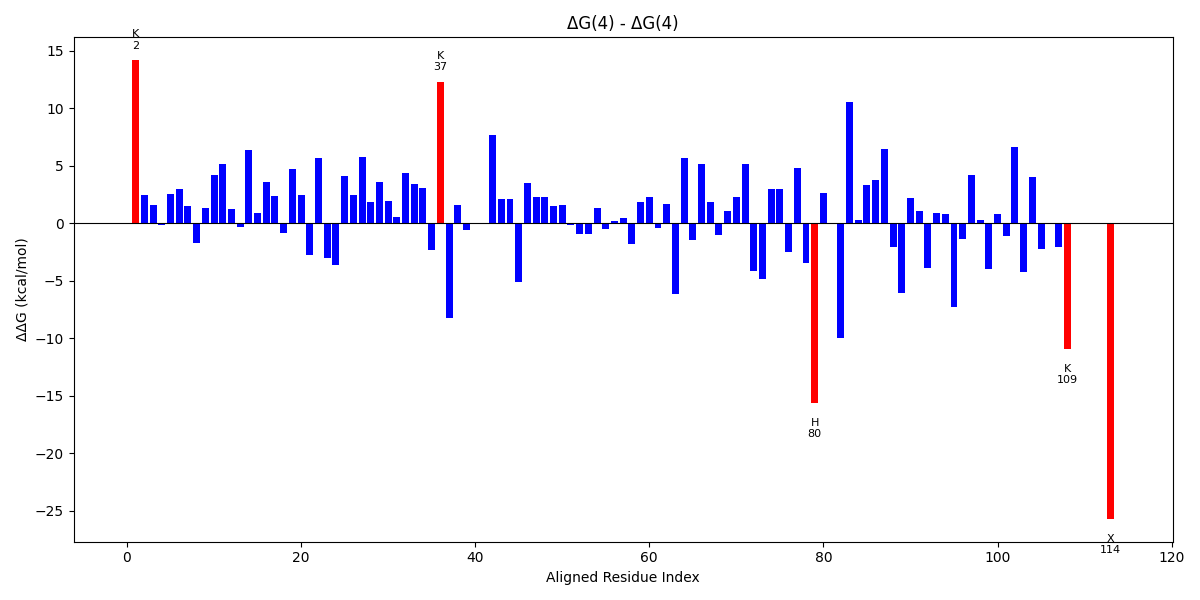
| aln_pos | aa1 | aa2 | E1 | E2 | dEdiff |
|---|---|---|---|---|---|
| 1 | - | M | - | 8.81 | - |
| 2 | K | K | 13.13 | -1.03 | 14.16 |
| 3 | D | D | 2.68 | 0.25 | 2.43 |
| 4 | R | R | 5.42 | 3.87 | 1.55 |
| 5 | K | K | 0.77 | 0.95 | -0.18 |
| 6 | I | I | 4.12 | 1.59 | 2.54 |
| 7 | L | L | -1.09 | -4.05 | 2.96 |
| 8 | N | N | -0.21 | -1.68 | 1.47 |
| 9 | E | E | 0.99 | 2.72 | -1.72 |
| 10 | I | I | -1.09 | -2.4 | 1.3 |
| 11 | L | L | 0.81 | -3.4 | 4.21 |
| 12 | S | S | 4.76 | -0.39 | 5.15 |
| 13 | N | N | 3.35 | 2.12 | 1.23 |
| 14 | T | T | -0.78 | -0.49 | -0.29 |
| 15 | I | I | 2.56 | -3.83 | 6.39 |
| 16 | N | N | 1.75 | 0.9 | 0.84 |
| 17 | E | E | 3.68 | 0.06 | 3.62 |
| 18 | L | L | -3.34 | -5.68 | 2.34 |
| 19 | N | N | -0.85 | 0.03 | -0.88 |
| 20 | L | L | 0.85 | -3.84 | 4.69 |
| 21 | N | N | 1.84 | -0.58 | 2.42 |
| 22 | D | D | -0.46 | 2.31 | -2.77 |
| 23 | K | K | 5.81 | 0.18 | 5.63 |
| 24 | K | K | -0.46 | 2.57 | -3.03 |
| 25 | A | A | -2.77 | 0.88 | -3.65 |
| 26 | N | N | 6.5 | 2.41 | 4.09 |
| 27 | I | I | -1.11 | -3.59 | 2.49 |
| 28 | K | K | 6.24 | 0.51 | 5.73 |
| 29 | I | I | -1.42 | -3.3 | 1.88 |
| 30 | K | K | 5.84 | 2.28 | 3.55 |
| 31 | I | I | 0.06 | -1.86 | 1.91 |
| 32 | K | K | 2.77 | 2.25 | 0.52 |
| 33 | P | P | 0.74 | -3.58 | 4.32 |
| 34 | L | L | -0.12 | -3.5 | 3.38 |
| 35 | K | K | 8.6 | 5.56 | 3.05 |
| 36 | R | R | 8.2 | 10.57 | -2.37 |
| 37 | K | K | 15.46 | 3.16 | 12.3 |
| 38 | I | I | 2.16 | 10.44 | -8.28 |
| 39 | A | A | -0.82 | -2.39 | 1.56 |
| 40 | S | S | -2.28 | -1.68 | -0.61 |
| 41 | I | I | -1.99 | -1.93 | -0.07 |
| 42 | S | S | 0.08 | 0.09 | -0.01 |
| 43 | L | L | 7.11 | -0.52 | 7.63 |
| 44 | T | T | 4.32 | 2.23 | 2.09 |
| 45 | N | N | 4.71 | 2.58 | 2.14 |
| 46 | K | K | 1.78 | 6.87 | -5.09 |
| 47 | T | T | 0.03 | -3.42 | 3.45 |
| 48 | I | I | -2.65 | -4.9 | 2.25 |
| 49 | Y | Y | -0.68 | -2.93 | 2.25 |
| 50 | I | I | -2.16 | -3.67 | 1.52 |
| 51 | N | N | -3.63 | -5.2 | 1.58 |
| 52 | K | K | 1.34 | 1.51 | -0.18 |
| 53 | N | N | -0.71 | 0.25 | -0.96 |
| 54 | I | I | 0.4 | 1.32 | -0.92 |
| 55 | L | L | 0.3 | -1.02 | 1.32 |
| 56 | P | P | -2.45 | -1.92 | -0.54 |
| 57 | Y | Y | 0.5 | 0.27 | 0.23 |
| 58 | L | L | -3.11 | -3.55 | 0.44 |
| 59 | S | S | -2.53 | -0.71 | -1.82 |
| 60 | D | D | 0.1 | -1.78 | 1.88 |
| 61 | E | E | 3.6 | 1.32 | 2.28 |
| 62 | E | E | -1.55 | -1.15 | -0.4 |
| 63 | I | I | -1.52 | -3.15 | 1.63 |
| 64 | R | R | -1.7 | 4.43 | -6.13 |
| 65 | F | F | 0.86 | -4.81 | 5.67 |
| 66 | I | I | 0.33 | 1.78 | -1.45 |
| 67 | L | L | 0.18 | -4.96 | 5.14 |
| 68 | A | A | -1.94 | -3.79 | 1.85 |
| 69 | H | H | 6.26 | 7.3 | -1.04 |
| 70 | E | E | -0.1 | -1.17 | 1.07 |
| 71 | L | L | -4.03 | -6.28 | 2.25 |
| 72 | L | L | 0.04 | -5.08 | 5.13 |
| 73 | H | H | 6.02 | 10.15 | -4.13 |
| 74 | L | L | -2.83 | 2.02 | -4.85 |
| 75 | K | K | -1.26 | -4.24 | 2.98 |
| 76 | Y | Y | -0.02 | -2.98 | 2.96 |
| 77 | G | G | -1.43 | 1.08 | -2.5 |
| 78 | K | K | 7.81 | 3.04 | 4.77 |
| 79 | Y | Y | -0.81 | 2.68 | -3.49 |
| 80 | H | H | 1.6 | 17.2 | -15.6 |
| 81 | I | I | 5.01 | 2.41 | 2.6 |
| 82 | N | N | 3.72 | 3.74 | -0.02 |
| 83 | E | E | 4.17 | 14.18 | -10.01 |
| 84 | F | F | 7.98 | -2.52 | 10.5 |
| 85 | E | E | 10.98 | 10.66 | 0.31 |
| 86 | E | E | 2.18 | -1.14 | 3.32 |
| 87 | E | E | 5.83 | 2.08 | 3.75 |
| 88 | L | L | 4.55 | -1.93 | 6.48 |
| 89 | L | L | 0.19 | 2.22 | -2.03 |
| 90 | F | F | -4.43 | 1.64 | -6.07 |
| 91 | L | L | 0.08 | -2.15 | 2.22 |
| 92 | F | F | -1.34 | -2.41 | 1.07 |
| 93 | P | P | -3.03 | 0.87 | -3.89 |
| 94 | N | N | -1.01 | -1.91 | 0.9 |
| 95 | K | K | 3.04 | 2.23 | 0.81 |
| 96 | E | E | 1.01 | 8.26 | -7.26 |
| 97 | A | A | -1.63 | -0.24 | -1.39 |
| 98 | I | I | 1.55 | -2.68 | 4.22 |
| 99 | L | L | -1.52 | -1.81 | 0.3 |
| 100 | I | I | 0.79 | 4.75 | -3.96 |
| 101 | N | N | 1.51 | 0.75 | 0.77 |
| 102 | L | L | 3.01 | 4.12 | -1.11 |
| 103 | I | I | 9.44 | 2.84 | 6.6 |
| 104 | N | N | 0.07 | 4.31 | -4.24 |
| 105 | K | K | 5.43 | 1.43 | 3.99 |
| 106 | L | L | 1.97 | 4.24 | -2.26 |
| 107 | H | H | 3.15 | 3.11 | 0.03 |
| 108 | Q | Q | 7.29 | 9.35 | -2.07 |
| 109 | K | K | 3.89 | 14.82 | -10.93 |
| 110 | X | - | 5.89 | - | - |
| 111 | X | - | -1.08 | - | - |
| 112 | X | - | 2.63 | - | - |
| 113 | X | - | -0.06 | - | - |
| 114 | X | X | 1.43 | 27.12 | -25.7 |
| 115 | X | X | 1.98 | 2.02 | -0.04 |